Welcome

A large number of research groups world-wide are using urine for measuring their marker of interest for kidney, bladder and prostate diseases. Some of these studies concern RNA transcripts that indicate the presence or progression of disease. Examples include the PCA3 and TMPRSS2-ERG transcripts for prostate cancer, the FGFR3 mutations for bladder cancer and presence of viral and bacterial sequences indicating infections. Besides cancer and infection, also other diseases are expected to be detectable at the DNA and RNA level, including: kidney stones, kidney transplantation, congenital kidney/bladder problems, BPH, cysts, and more. The ongoing efforts show that urine is an excellent source of relevant biomarkers: urine is the ultimate ‘liquid biopsy’ for urinary tract diseases.
The developments of Next Generation Sequencing (NGS) are now at the level that we can sequence all transcripts and DNA present in tissues, but also in serum and urine. The RNA in body fluids is mainly present in small extracellular vesicles, often referred to as exosomes when up to ~200 nm in diameter. Also larger vesicles are often observed and named microvesicles or apoptotic vesicles. These vesicles are secreted by most cell types and are filled with the content of the cell, including their proteins, RNAs and DNA. Also cancer cells secrete these vesicles, which end up in the urine and blood. Importantly, the vesicles protect the RNA from degradation. Since diseased cells (including cancer cells) express specific RNAs (such as the TMPRSS2-ERG or mutated FGFR3 transcripts), presence and status of disease can be measured by RNA sequencing of exosomal RNAs in body fluids.
For urine, full transcriptome and DNA sequencing has not been performed at any large scale yet. With the NGS promise that it will become affordable to sequence urinary transcripts in a clinical setting, it is timely to develop the protocols and initiate the creation of a database of urine transcriptomes for healthy and sick children, women and men: The Urinome Project. The Urinome database should become a standard to discover and validate urinary tract RNA and DNA markers.
The ultimate goal of the Urinome Project is to discover, validate and implement urinary markers for urinary tract diseases by create a database with fully sequenced genomes and transcriptomes of a large collection of urine samples. Any disease (infection, stones, cancer, chronic and congenital diseases, transplantation, etc.) of the kidney, bladder and prostate will be included.


Project Description
The Urinome Project consists of two phases:
In the feasibility phase, we will develop the standard operating procedures (SOPs) for urine collection, processing, storage, RNA extraction and sequencing (small and long RNAs, combined). We will determine which fraction of urine (whole urine, sediment, supernatant, extracellular vesicles) is most suited for sequencing.
The bioinformatics pipelines and database structure will be set up and the minimal required information on the SOPs, quality control and patient information determined for data to enter the Urinome database. Medical ethical approvals, data security and privacy concerns will be addressed.
In the execution phase, participants perform the DNA and RNA sequencing. The database will be filled with patient and sequencing data and upon approval, made available to the research community.
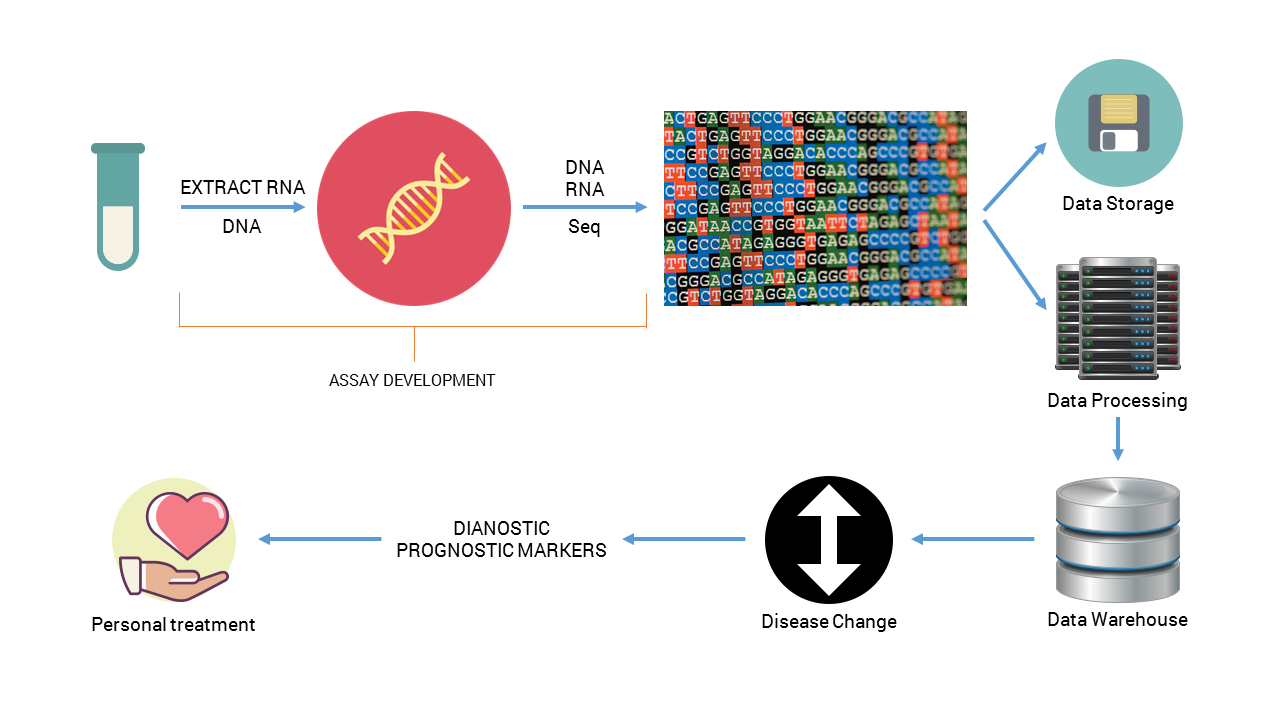
Project workflow. Upon DNA and RNA extraction and subsequent sequencing, data will be stored and processed to identify characteristic disease-associated changes that are markers for better and personalized treatment.


Funding
Funding has been provided by kind donations through the SUWO and the Erasmus MC Vriendenfonds.


News & Publications


Contact
Details
Professor Experimental Urological Oncology
Address:
Josephine Nefkens Institute, Room Be362a
Erasmus MC
P.O. box 2040
3000 CA Rotterdam, The Netherlands
Phone: +31-10-704-3672
E-mail: g.jenster@erasmusmc.nl